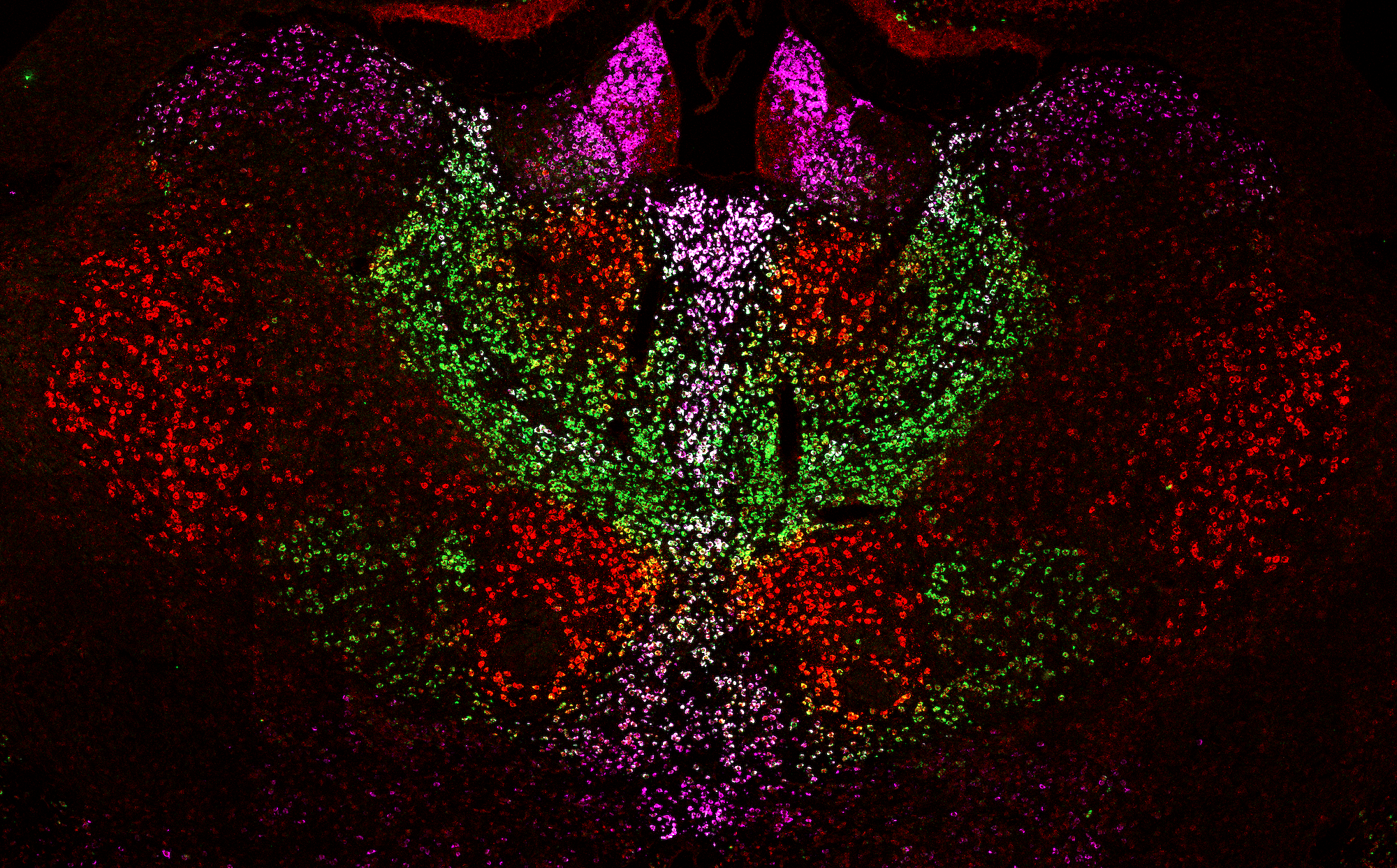
Thalamoseq
Deep sequencing of the cellular transcriptome has provided unprecedented access to the molecular diversity of neurons. However, it remains a challenge to make sense of the observed molecular diversity of neurons - including the challenge of relating molecular diversity to diversity in projection patterns, dendritic morphology, cellular biophysics, etc. To address this question a joint graduate student in our lab and the Hantman lab began a project to assess the molecular diversity of the mouse thalamus. We sought to use these approaches to both understand the logic by which the ~30 thalamic nuclei are organized as well as to provide molecular entry points that allow for precise manipulation of thalamic function. This developed into a project that drew upon diverse expertise within Janelia and in collaboration with Sacha Nelson at Brandeis. This effort produced a large dataset that describes molecular and anatomical diversity of major thalamic nuclei. The data that is publicly available at thalamoseq.janelia.org and the work is described in a recent paper in Nature Neuroscience.
This work was led by Adam Hantman's lab in collaboration with Sacha Nelson's lab and Quantitative Genomics in addition to the Neuroseq and MouseLight Janelia Project Teams, and Scientific Computing made the online resources for browsing the data.
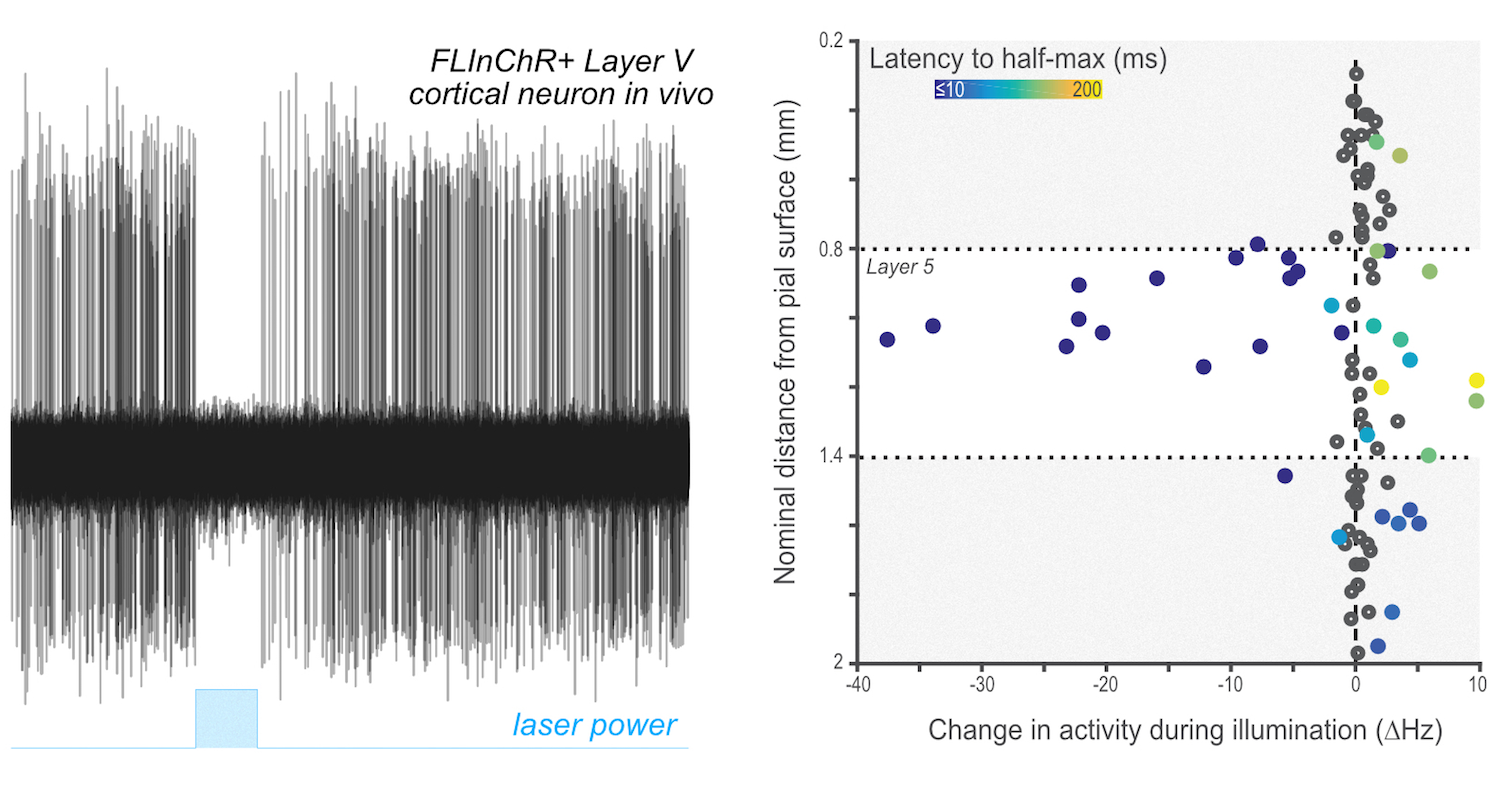
flinchr
Optogenetics provides an exceptional and powerful toolkit for the exploration of brain function by allowing precise manipulation of individual cell types and circuits. Remarkably efficient tools exist for the stimulation of individual neurons discovered via mining of diverse genomes; however, while there are a number of tools that allow for inhibition - the result of both mining extant genomes and substantial directed mutagenesis - such tools are generally less robust and reliable in practice for many neuroscience applications. This suggests that alternate methods of diversifying the toolkit of available optogenetics reagents is required. While site directed mutagenesis has been a proven method we sought to exploit another method of diversification in evolution - recombination. Recombination is a process by which large coding regions of distinct genes may be combined to yield hybrid proteins of (potentially) novel function. In collaboration with Alla Karpova's lab several years ago we sought to begin to explore how to produce hybrid reagents through the modular combination of existing reagents and targeting domains. In one of first products of this collaboration we described the production of FLInChR (Full length inversion of channelrhodopsin) through the combination of a transmembrane domain on to the N-terminus of channelrhodopsin (ChR). This produced a topological inversion of ChR that, due to its pump-like properties, produced a dramatic alteration in function: it converted a neuronal activator into a potent neuronal silencer as described in our recent paper and summarized in a few places. Example data showing complete silencing of a relatively difficult to silence neuronal cell type (large layer 5 pyramidal cells in neocortex) is shown at right.
Through ongoing efforts we continue to explore this method of diversification of our toolkit for systems neuroscience and hope to produce additional useful resources. If you would like to try this inhibitor the plasmid is available at AddGene.
Nanshe
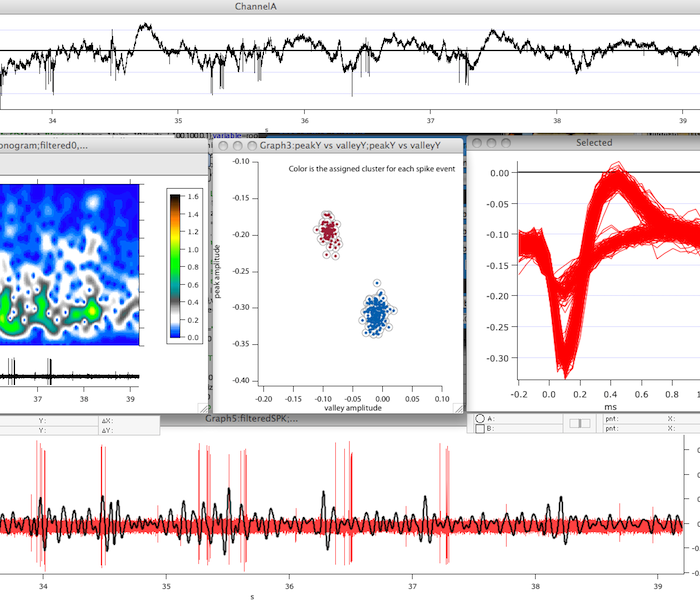
Our workflow for automated analysis of calcium imaging videos is written in Python and is compatible with both version 2 and 3. Leveraging the power and interactivity of Jupyter notebooks, we have developed a solution for rigid body registration of images, automated ROI detection, and spike extraction by combining a variety of open source tools. The workflow manages conversion of tif stacks into HDF5 data files for efficient processing and distribution across powerful compute clusters. In addition, by implementing the workflow in a Jupyter notebook intermediate analyses can be visualized in embedded, interactive graphs (as seen at left). The workflow provides extensibility by exposing key steps in the workflow with a simple API to allow custom algorithms for analysis and pre-processing to be rapidly incorporated.
This grew out of a collaborative visitor project with Ferran Diego in Fred Hamprecht's group and is a modular workflow that can implement their dictionary learning based algorithm for automatic ROI generation. In this version of the workflow we expose scikit-learn with a simple API and allowing users to flexibly apply a range of algorithms for learning basis images (or any other preferred approach).
Click (+shift) on ROI to view timeseries data (example from neurofinder data).
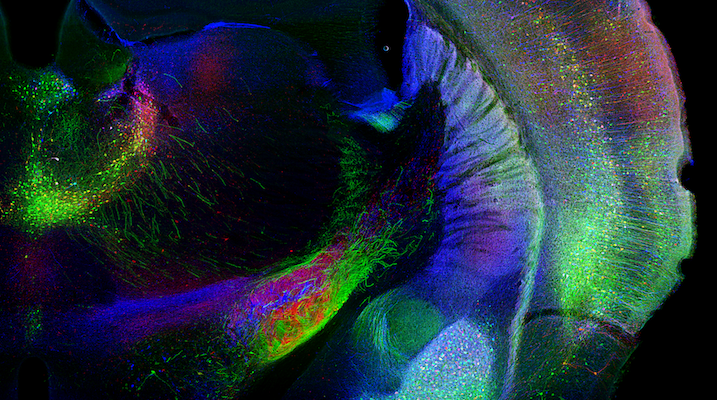
retro-AAV
Efficient retrograde access to projection neurons with the aim of delivering sensors and effectors constitutes an important future avenue for circuit dissection. Viral vectors are powerful gene delivery vehicles for the nervous system, but all available viral tools suffer from inefficient retrograde transport. Using directed molecular evolution together with our collaborators we engineered potent retrograde functionality into the capsid of AAV — a vector that already shows particular promise in neuroscience research. The newly evolved AAV2-retro variant permits robust retrograde access to projection neurons with efficiency comparable to classical synthetic retrograde labeling reagents, and supports high-level payload expression adequate for functional circuit interrogation.
Plasmid is available at AddGene.
We have set up a large-scale production of the following three basic tester preps at the Janelia facility: rAAV2-retro CAG-tdtomato, rAAV2-retro FLEX-tdtomato and rAAV2-retro Syn-Cre.
These preps should be available within about a month. If you are unsure about the results you obtain with preps from any other sources, you will be able to request a free tester aliquot to use as a standard.
To obtain a tester prep, please send a request to Janelia Virus Services or contact my lab.

Skilled movements & decisions
We have argued that the measurement of kinematic parameters of goal-directed movements is important for understanding the function of basal ganglia circuits that control purposive behavior.
Recently, we have developed a behavioral task in which mice displace a spring-loaded 2D joystick past a hidden threshold to receive reward. Assuming a roughly constant transfer of work from the limbs to the joystick effort is related to the velocity of joystick movement.
By using a continuous analog signal proportional to movement kinematics we can study how effort is traded off against reward, continuous neural representations of movement kinematics, as well as performing rapid (kHz), closed-loop feedback.
The system we developed has 3 key parts:
- a hall-effect sensor based, spring loaded joystick like this one;
- a mechanical system for stable head fixation. We like RIVETS;
- an embedded controller with fast, good A/D conversion to allow for rapid closed-loop feedback. We like the behavioral control system III.
You can contact us with specific questions or inquiries about the software we use to interact with the BCS if needed; however, there are many solutions available.
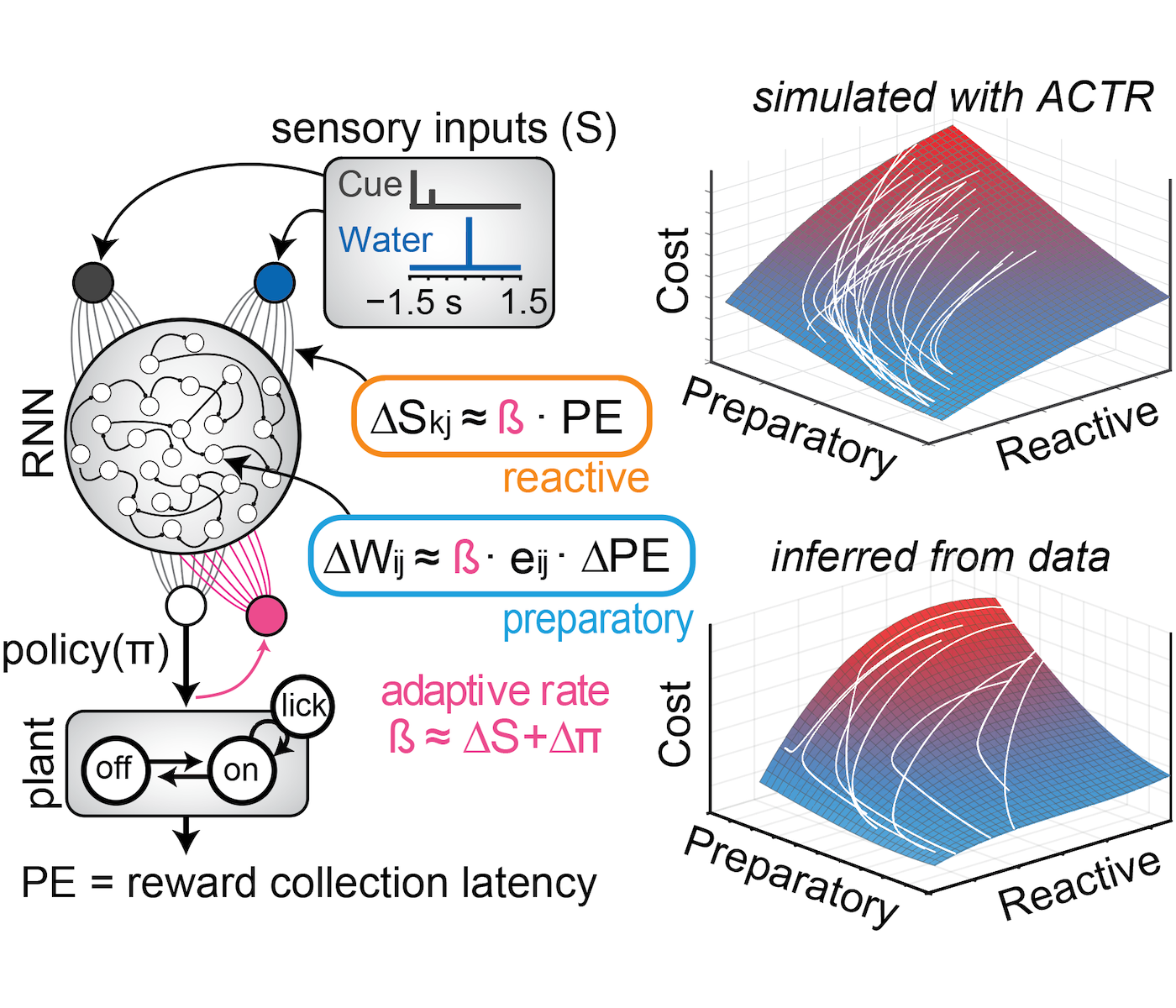
RNN-based reinforcement learning
Source code describing the reinforcement learning model developed in Coddington, Lindo & Dudman, 2023.

RIVETS
A number of recent studies have provided compelling demonstrations that mice and rats can be trained to perform a variety of tasks while head restrained. The independent development of this technique by a number of labs has led to a variety of solutions, but these solutions are often expensive or impede rapid development and modification. To address these issues, we developed a flexible single platform for electrophysiology and imaging both ex vivo and in vivo. Our design facilitates the rapid conversion of a single physiology and imaging system between preparations and is used by many labs at Janelia Research Campus and sold by Ronal Tool.

BCS
The Behavioral Control System is designed to work with behavioral assay rigs that require data acquisition and digital processing. The developed system has the capability to drive switches and solenoids as well as control and monitor beam breaks. It also has a pulse generator with analog output levels. The main processing controller is an Arduino Mega board with an additional microcontroller with Teensy breakout board. The BNC input/output connectors have different options the user can select depending on research needs (analog input/output, digital input/output etc.).
Code for running the BCS III board.
Corduroy
We have developed a complete data acquisition and analysis suite of functions written for Igor Pro and uses National Instruments hardware. Both continuous acquistion and episodic acquisition are supported with evolving support for multichannel recordings. If you are interested in receiving a distribution copy head over to our GitHub repo. Full disclosure: the manual at the moment is abysmal so it is really only suitable for a user with lots of experience writing code in IgorPro. We hope to change this.
We typically use the PCIe-6259 board.

Cheap Track
As part of our work to characterize mice with a variety of manipulations or neural signaling (both acute and chronic) we found ourselves often needing to characterize basic features of locomotion. We wanted a cheap and easy solution. The result was a small piece of code written in JAVA using some open source libraries to extract position of a moving "blob" in real-time. If you find a need for such a piece of code you can feel free to download it and try it out. We have tested the software with this and this camera, but it will work with any USB camera that Quicktime can recognize.
Stellate
Source code for a minimal version of the model of a stochastic stellate neuron developed in Dudman & Nolan, 2009. The code is a brute force stimulation of a number of ion channels with stochastic gating written in Matlab. Model is hand tuned to stellate neuron physiology and is able to replicate a number of features in the basic physiology (e.g. Nolan et al., 2007).